
Autoplot method for discharge rating curve tournament
Source:R/tournament_methods.R
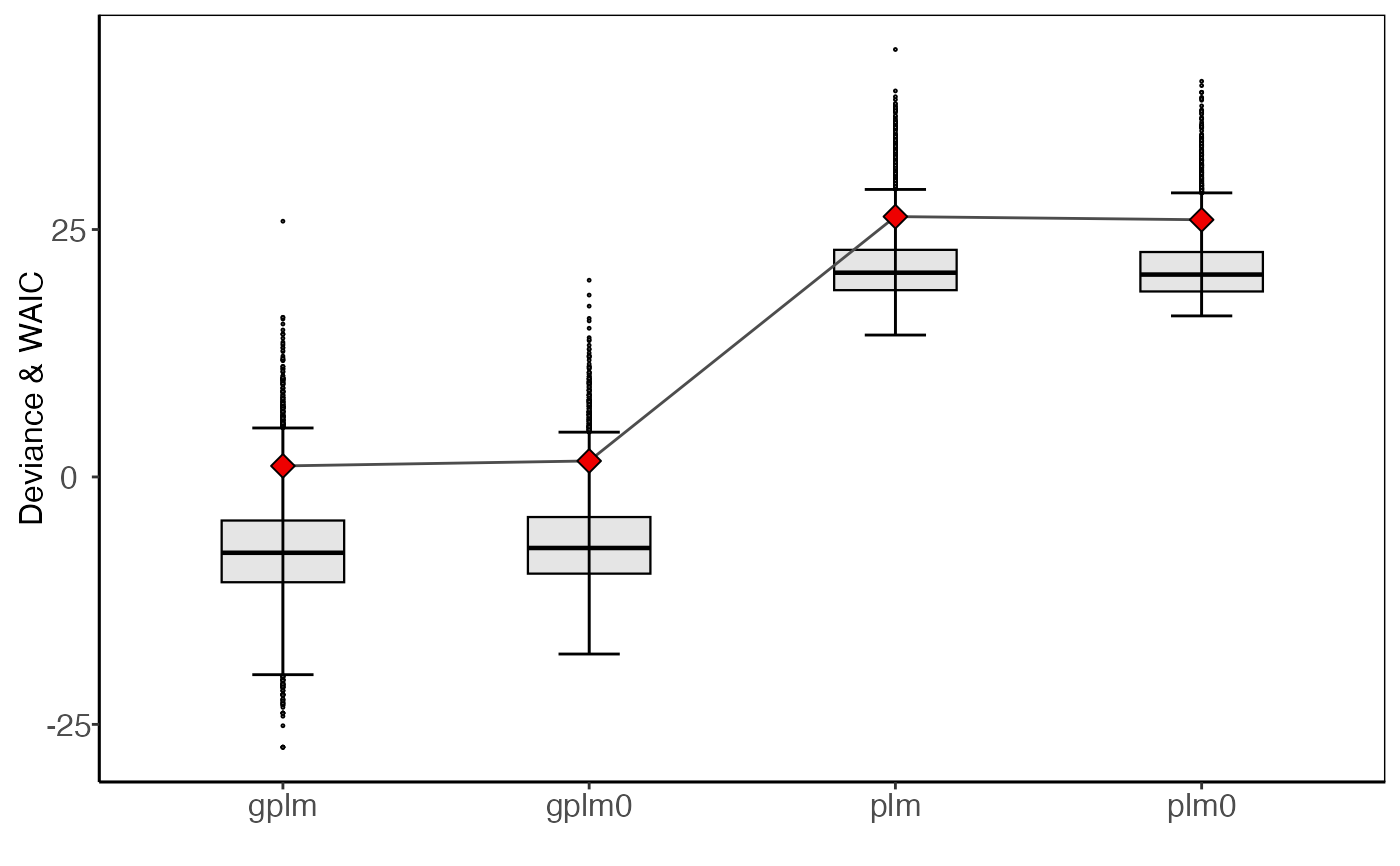
autoplot.tournament.RdCompare the four discharge rating curves from the tournament object in different ways
Usage
# S3 method for class 'tournament'
autoplot(object, type = "boxplot", ...)See also
tournament to run a discharge rating curve tournament and summary.tournament for summaries.
Examples
# \donttest{
library(ggplot2)
data(krokfors)
set.seed(1)
t_obj <- tournament(formula = Q ~ W, data = krokfors, num_cores = 2)
#> Running tournament [ ] 0%
#>
#> Progress:
#> Initializing Metropolis MCMC algorithm...
#> Multiprocess sampling (4 chains in 2 jobs) ...
#>
#> MCMC sampling completed!
#>
#> Diagnostics:
#> Acceptance rate: 25.33%.
#> ✔ All chains have mixed well (Rhat < 1.1).
#> ✔ Effective sample sizes sufficient (eff_n_samples > 400).
#>
#> ✔ gplm finished [============ ] 25%
#>
#> Progress:
#> Initializing Metropolis MCMC algorithm...
#> Multiprocess sampling (4 chains in 2 jobs) ...
#>
#> MCMC sampling completed!
#>
#> Diagnostics:
#> Acceptance rate: 31.14%.
#> ✔ All chains have mixed well (Rhat < 1.1).
#> ✔ Effective sample sizes sufficient (eff_n_samples > 400).
#>
#> ✔ gplm0 finished [======================== ] 50%
#>
#> Progress:
#> Initializing Metropolis MCMC algorithm...
#> Multiprocess sampling (4 chains in 2 jobs) ...
#>
#> MCMC sampling completed!
#>
#> Diagnostics:
#> Acceptance rate: 25.66%.
#> ✔ All chains have mixed well (Rhat < 1.1).
#> ✔ Effective sample sizes sufficient (eff_n_samples > 400).
#>
#> ✔ plm finished [==================================== ] 75%
#>
#> Progress:
#> Initializing Metropolis MCMC algorithm...
#> Multiprocess sampling (4 chains in 2 jobs) ...
#>
#> MCMC sampling completed!
#>
#> Diagnostics:
#> Acceptance rate: 36.04%.
#> ✔ All chains have mixed well (Rhat < 1.1).
#> ✔ Effective sample sizes sufficient (eff_n_samples > 400).
#>
#> ✔ plm0 finished [================================================] 100%
autoplot(t_obj)
 # }
# }